In [1]: import pandas as pd
-
Titanic data
This tutorial uses the Titanic data set, stored as CSV. The data consists of the following data columns:
PassengerId: Id of every passenger.
Survived: This feature have value 0 and 1. 0 for not survived and 1 for survived.
Pclass: There are 3 classes: Class 1, Class 2 and Class 3.
Name: Name of passenger.
Sex: Gender of passenger.
Age: Age of passenger.
SibSp: Indication that passenger have siblings and spouse.
Parch: Whether a passenger is alone or have family.
Ticket: Ticket number of passenger.
Fare: Indicating the fare.
Cabin: The cabin of passenger.
Embarked: The embarked category.
In [2]: titanic = pd.read_csv("data/titanic.csv") In [3]: titanic.head() Out[3]: PassengerId Survived Pclass Name ... Ticket Fare Cabin Embarked 0 1 0 3 Braund, Mr. Owen Harris ... A/5 21171 7.2500 NaN S 1 2 1 1 Cumings, Mrs. John Bradley (Florence Briggs Th... ... PC 17599 71.2833 C85 C 2 3 1 3 Heikkinen, Miss. Laina ... STON/O2. 3101282 7.9250 NaN S 3 4 1 1 Futrelle, Mrs. Jacques Heath (Lily May Peel) ... 113803 53.1000 C123 S 4 5 0 3 Allen, Mr. William Henry ... 373450 8.0500 NaN S [5 rows x 12 columns]
-
Air quality data
This tutorial uses air quality data about \(NO_2\) and Particulate matter less than 2.5 micrometers, made available by openaq and using the py-openaq package. The
air_quality_long.csvdata set provides \(NO_2\) and \(PM_{25}\) values for the measurement stations FR04014, BETR801 and London Westminster in respectively Paris, Antwerp and London.The air-quality data set has the following columns:
city: city where the sensor is used, either Paris, Antwerp or London
country: country where the sensor is used, either FR, BE or GB
location: the id of the sensor, either FR04014, BETR801 or London Westminster
parameter: the parameter measured by the sensor, either \(NO_2\) or Particulate matter
value: the measured value
unit: the unit of the measured parameter, in this case ‘µg/m³’
and the index of the
DataFrameisdatetime, the datetime of the measurement.To raw dataNote
The air-quality data is provided in a so-called long format data representation with each observation on a separate row and each variable a separate column of the data table. The long/narrow format is also known as the tidy data format.
In [4]: air_quality = pd.read_csv( ...: "data/air_quality_long.csv", index_col="date.utc", parse_dates=True ...: ) ...: In [5]: air_quality.head() Out[5]: city country location parameter value unit date.utc 2019-06-18 06:00:00+00:00 Antwerpen BE BETR801 pm25 18.0 µg/m³ 2019-06-17 08:00:00+00:00 Antwerpen BE BETR801 pm25 6.5 µg/m³ 2019-06-17 07:00:00+00:00 Antwerpen BE BETR801 pm25 18.5 µg/m³ 2019-06-17 06:00:00+00:00 Antwerpen BE BETR801 pm25 16.0 µg/m³ 2019-06-17 05:00:00+00:00 Antwerpen BE BETR801 pm25 7.5 µg/m³
How to reshape the layout of tables?¶
Sort table rows¶
I want to sort the Titanic data according to the age of the passengers.
In [6]: titanic.sort_values(by="Age").head() Out[6]: PassengerId Survived Pclass Name Sex Age SibSp Parch Ticket Fare Cabin Embarked 803 804 1 3 Thomas, Master. Assad Alexander male 0.42 0 1 2625 8.5167 NaN C 755 756 1 2 Hamalainen, Master. Viljo male 0.67 1 1 250649 14.5000 NaN S 644 645 1 3 Baclini, Miss. Eugenie female 0.75 2 1 2666 19.2583 NaN C 469 470 1 3 Baclini, Miss. Helene Barbara female 0.75 2 1 2666 19.2583 NaN C 78 79 1 2 Caldwell, Master. Alden Gates male 0.83 0 2 248738 29.0000 NaN S
I want to sort the Titanic data according to the cabin class and age in descending order.
In [7]: titanic.sort_values(by=['Pclass', 'Age'], ascending=False).head() Out[7]: PassengerId Survived Pclass Name Sex Age SibSp Parch Ticket Fare Cabin Embarked 851 852 0 3 Svensson, Mr. Johan male 74.0 0 0 347060 7.7750 NaN S 116 117 0 3 Connors, Mr. Patrick male 70.5 0 0 370369 7.7500 NaN Q 280 281 0 3 Duane, Mr. Frank male 65.0 0 0 336439 7.7500 NaN Q 483 484 1 3 Turkula, Mrs. (Hedwig) female 63.0 0 0 4134 9.5875 NaN S 326 327 0 3 Nysveen, Mr. Johan Hansen male 61.0 0 0 345364 6.2375 NaN S
With
Series.sort_values(), the rows in the table are sorted according to the defined column(s). The index will follow the row order.
More details about sorting of tables is provided in the using guide section on sorting data.
Long to wide table format¶
Let’s use a small subset of the air quality data set. We focus on
\(NO_2\) data and only use the first two measurements of each
location (i.e. the head of each group). The subset of data will be
called no2_subset
# filter for no2 data only
In [8]: no2 = air_quality[air_quality["parameter"] == "no2"]
# use 2 measurements (head) for each location (groupby)
In [9]: no2_subset = no2.sort_index().groupby(["location"]).head(2)
In [10]: no2_subset
Out[10]:
city country location parameter value unit
date.utc
2019-04-09 01:00:00+00:00 Antwerpen BE BETR801 no2 22.5 µg/m³
2019-04-09 01:00:00+00:00 Paris FR FR04014 no2 24.4 µg/m³
2019-04-09 02:00:00+00:00 London GB London Westminster no2 67.0 µg/m³
2019-04-09 02:00:00+00:00 Antwerpen BE BETR801 no2 53.5 µg/m³
2019-04-09 02:00:00+00:00 Paris FR FR04014 no2 27.4 µg/m³
2019-04-09 03:00:00+00:00 London GB London Westminster no2 67.0 µg/m³
I want the values for the three stations as separate columns next to each other
In [11]: no2_subset.pivot(columns="location", values="value") Out[11]: location BETR801 FR04014 London Westminster date.utc 2019-04-09 01:00:00+00:00 22.5 24.4 NaN 2019-04-09 02:00:00+00:00 53.5 27.4 67.0 2019-04-09 03:00:00+00:00 NaN NaN 67.0
The
pivot()function is purely reshaping of the data: a single value for each index/column combination is required.
As pandas support plotting of multiple columns (see plotting tutorial) out of the box, the conversion from long to wide table format enables the plotting of the different time series at the same time:
In [12]: no2.head()
Out[12]:
city country location parameter value unit
date.utc
2019-06-21 00:00:00+00:00 Paris FR FR04014 no2 20.0 µg/m³
2019-06-20 23:00:00+00:00 Paris FR FR04014 no2 21.8 µg/m³
2019-06-20 22:00:00+00:00 Paris FR FR04014 no2 26.5 µg/m³
2019-06-20 21:00:00+00:00 Paris FR FR04014 no2 24.9 µg/m³
2019-06-20 20:00:00+00:00 Paris FR FR04014 no2 21.4 µg/m³
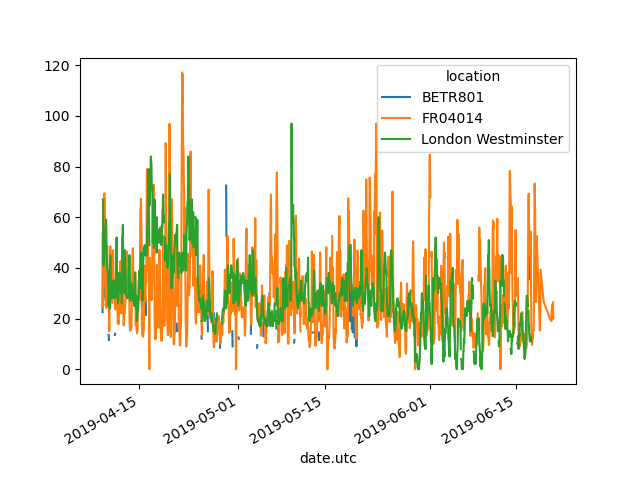
In [13]: no2.pivot(columns="location", values="value").plot()
Out[13]: <AxesSubplot:xlabel='date.utc'>
Note
When the index parameter is not defined, the existing
index (row labels) is used.
For more information about pivot(), see the user guide section on pivoting DataFrame objects.
Pivot table¶
I want the mean concentrations for \(NO_2\) and \(PM_{2.5}\) in each of the stations in table form
In [14]: air_quality.pivot_table( ....: values="value", index="location", columns="parameter", aggfunc="mean" ....: ) ....: Out[14]: parameter no2 pm25 location BETR801 26.950920 23.169492 FR04014 29.374284 NaN London Westminster 29.740050 13.443568
In the case of
pivot(), the data is only rearranged. When multiple values need to be aggregated (in this specific case, the values on different time steps)pivot_table()can be used, providing an aggregation function (e.g. mean) on how to combine these values.
Pivot table is a well known concept in spreadsheet software. When
interested in summary columns for each variable separately as well, put
the margin parameter to True:
In [15]: air_quality.pivot_table(
....: values="value",
....: index="location",
....: columns="parameter",
....: aggfunc="mean",
....: margins=True,
....: )
....:
Out[15]:
parameter no2 pm25 All
location
BETR801 26.950920 23.169492 24.982353
FR04014 29.374284 NaN 29.374284
London Westminster 29.740050 13.443568 21.491708
All 29.430316 14.386849 24.222743
For more information about pivot_table(), see the user guide section on pivot tables.
Note
In case you are wondering, pivot_table() is indeed directly linked
to groupby(). The same result can be derived by grouping on both
parameter and location:
air_quality.groupby(["parameter", "location"]).mean()
Have a look at groupby() in combination with unstack() at the user guide section on combining stats and groupby.
Wide to long format¶
Starting again from the wide format table created in the previous section:
In [16]: no2_pivoted = no2.pivot(columns="location", values="value").reset_index()
In [17]: no2_pivoted.head()
Out[17]:
location date.utc BETR801 FR04014 London Westminster
0 2019-04-09 01:00:00+00:00 22.5 24.4 NaN
1 2019-04-09 02:00:00+00:00 53.5 27.4 67.0
2 2019-04-09 03:00:00+00:00 54.5 34.2 67.0
3 2019-04-09 04:00:00+00:00 34.5 48.5 41.0
4 2019-04-09 05:00:00+00:00 46.5 59.5 41.0
I want to collect all air quality \(NO_2\) measurements in a single column (long format)
In [18]: no_2 = no2_pivoted.melt(id_vars="date.utc") In [19]: no_2.head() Out[19]: date.utc location value 0 2019-04-09 01:00:00+00:00 BETR801 22.5 1 2019-04-09 02:00:00+00:00 BETR801 53.5 2 2019-04-09 03:00:00+00:00 BETR801 54.5 3 2019-04-09 04:00:00+00:00 BETR801 34.5 4 2019-04-09 05:00:00+00:00 BETR801 46.5
The
pandas.melt()method on aDataFrameconverts the data table from wide format to long format. The column headers become the variable names in a newly created column.
The solution is the short version on how to apply pandas.melt(). The method
will melt all columns NOT mentioned in id_vars together into two
columns: A column with the column header names and a column with the
values itself. The latter column gets by default the name value.
The pandas.melt() method can be defined in more detail:
In [20]: no_2 = no2_pivoted.melt(
....: id_vars="date.utc",
....: value_vars=["BETR801", "FR04014", "London Westminster"],
....: value_name="NO_2",
....: var_name="id_location",
....: )
....:
In [21]: no_2.head()
Out[21]:
date.utc id_location NO_2
0 2019-04-09 01:00:00+00:00 BETR801 22.5
1 2019-04-09 02:00:00+00:00 BETR801 53.5
2 2019-04-09 03:00:00+00:00 BETR801 54.5
3 2019-04-09 04:00:00+00:00 BETR801 34.5
4 2019-04-09 05:00:00+00:00 BETR801 46.5
The result in the same, but in more detail defined:
value_varsdefines explicitly which columns to melt togethervalue_nameprovides a custom column name for the values column instead of the default column namevaluevar_nameprovides a custom column name for the column collecting the column header names. Otherwise it takes the index name or a defaultvariable
Hence, the arguments value_name and var_name are just
user-defined names for the two generated columns. The columns to melt
are defined by id_vars and value_vars.
Conversion from wide to long format with pandas.melt() is explained in the user guide section on reshaping by melt.
REMEMBER
Sorting by one or more columns is supported by
sort_valuesThe
pivotfunction is purely restructuring of the data,pivot_tablesupports aggregationsThe reverse of
pivot(long to wide format) ismelt(wide to long format)
A full overview is available in the user guide on the pages about reshaping and pivoting.